Circadian clocks synchronize cellular metabolism to the diurnal light cycle. In humans, our biological clocks play important roles in our physiologies, including sleep, mental well-being, and disease states. Despite great progress in understanding the function of clock genes, the molecular mechanisms that govern underlying transcriptional feedback loops and their light entrainment are still not well understood.
In general, the clock consists of transcriptional-translational feedback loops wherein repressor proteins inhibit the transcriptional activators of their own genes and thus result in oscillatory gene expression and behavioral patterns. Light entrains the clock phase by stimulating photosensors that impinge directly on these components. Eukaryotic model systems such as Neurospora (filamentous fungi) and Drosophila (flies) are of particular interest, as they share similarities with higher metazoan clocks and have well defined photoreceptors and homologous molecular oscillators. These model systems also exemplify the general strategies used by higher organisms for light setting: transcription factor activation and repressor destruction.
A combination of crystallographic and spectroscopic approaches, along with the rational design and production of variant photosensors with perturbed photocycles, redox potentials, or conformational coupling, are currently being employed to understand activation mechanisms and molecular interactions with downstream targets.
Structure of full-length Drosophila cryptochrome
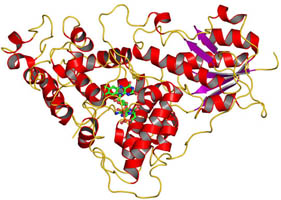

The cryptochrome/photolyase (CRY/PL) family of photoreceptors mediates adaptive responses to ultraviolet and blue light exposure in all kingdoms of life. Whereas PLs function predominantly in DNA repair of cyclobutane pyrimidine dimers (CPDs) and 6-4 photolesions caused by ultraviolet radiation, CRYs transduce signals important for growth, development, magnetosensitivity and circadian clocks. Despite these diverse functions, PLs/CRYs preserve a common structural fold, a dependence on flavin adenine dinucleotide (FAD) and an internal photoactivation mechanism. However, members of the CRY/PL family differ in the substrates recognized (protein or DNA), photochemical reactions catalysed and involvement of an antenna cofactor. It is largely unknown how the animal CRYs that regulate circadian rhythms act on their substrates. CRYs contain a variable carboxy-terminal tail that appends the conserved PL homology domain (PHD) and is important for function. We elucidated a 2.3-Å resolution crystal structure of Drosophila CRY with an intact C terminus. The C-terminal helix docks in the analogous groove that binds DNA substrates in PLs. Conserved Trp 536 juts into the CRY catalytic centre to mimic PL recognition of DNA photolesions. The FAD anionic semiquinone found in the crystals assumes a conformation to facilitate restructuring of the tail helix. These results help reconcile the diverse functions of the CRY/PL family by demonstrating how conserved protein architecture and photochemistry can be elaborated into a range of light-driven functions.
Zoltowski BD, Vaidya AT, Top D, Widom J, Young MW, Crane BR. Structure of full-length Drosophila cryptochrome. Nature 2011, 480, 396 – U156.
See Cornell Chronicle article.
Structure of a Light-Activated LOV Protein Dimer That Regulates Transcription
The filamentous fungus Neurospora crassa uses two blue-light sensors, white collar-1 (WC-1) and VIVID (VVD), both of which contain light, oxygen, or voltage (LOV) domains, to regulate its responses to light. WC-1 and WC-2 form a complex (WCC) to drive expression of light-induced genes, including vvd, whereas VVD tunes responses to light by directly interacting with and antagonizing the function of WCC. Vaidya et al. solved the crystal structure of the light-induced dimer of VVD and compared it to previously determined structures of the dark-state, monomeric protein. This analysis revealed that light not only induced a conformational change in the positioning of the N-terminal cap of one subunit of VVD but also triggered the opening of a binding pocket in the opposing subunit, into which the N-terminal cap could dock. Determining the structure explained the functional responses observed in vvd-deficient Neurospora that expressed variant VVD proteins and may inform mechanisms by which LOV-containing proteins interact.
Vaidya AT, Chen CH, Dunlap JC, Loros JJ, Crane BR. Structure of a light-activated LOV protein dimer that regulates transcription in Neurospora crassa. Sci Signal. 2011, 4.